
Modern DNA analysis
Critical biological samples were taken from the victims, from suspects and from the collected evidence. The appeals attorney petitioned the courts to have DNA testing performed on this case evidence. The state and the appeals attorneys agreed to examine the following evidence.
Samples from the victims and their clothes.
- Routine fingernail scraping from each victim that may or not have tissue from the perpetrator (each hand samples saved separately, three victims for six evidence samples).
- Ligatures from each of the three victims
- Tissue, skin or hair samples obtained from the ligatures of each victim.
- Swabs taken from each victim.
- Cuttings from blue jeans.
- Cuttings from blue pants.
Anomalous hairs taken from the victims.
- Two Caucasian hairs removed from Moore.
- Dyed hair removed from Moore.
- Hair found on Byers' body
- Hair on lower body, Byers
- Hair, perineum, Byers
- Two dark Caucasian hairs removed from Branch.
- Although all hairs from the ligatures were requested as mentioned above, specific hairs were also requested from the bindings Moore and Byers ligatures.
Anomalous hairs from clothing and fabrics in contact with the children and the crime scene.
- Negroid hair removed from white sheet 1.
- Hair from blue pants.
- Hair from Scout cap.
- Hair on tree stump (crime scene)
And, finally, other items.
- Bag of clothing, area of homicide.
- Hair from two knives from Richard Cummings.
- Hair from knife from Jason Crosby/Richard Appling.
- Wooden stick, crime scene.
Biological specimens were also obtained from 44 other suspects and family members for matches or exclusion. These samples can be tested by the modern DNA techniques of STR fingerprinting and mtDNA. The results can then compared to known samples from the children, the convicted and others to determine the source.
STR Fingerprinting
Along with the meaningful DNA that encodes for physical traits, we have a lot of "junk DNA," strings of repeating patterns and filler material. STR stands for short terminal repeating sections, and this area of DNA represents a pattern, 2 to 7 letters long that repeats itself many times. How many times varies between people. Any given STR sequence is about as good as HLA DQalpha technique for determining relatedness. Fortunately, there are such sequences on each chromosome, each with its own length. By testing nine of these sites on different chromosomes you get a one in a billion unique signature. Nine sites as standards are used by the military for paternity matters. Thirteen sites are commonly used for forensic tests and for the CODIS database.
To perform STR analysis, you divide up a sample for separate PCR reactions. Each yields a product of a specific length of DNA, the length determined by the number of repeats within the sequence. These are then run on a gel to determine the exact length of each sequence. Again a specific stepladder-like pattern of DNA pieces with different lengths can be compared to a source sample.
Presented below are six STR loci, using the analogies of sentences. The various sequences "yada_yada," "etc_etc_etc" are present in differing amount of copies among individuals.
Suspect A
Site_one_yada_yada_yada_yada_yada_yada_site_one_end (51 characters)
Site_two_oy_oy_oy_oy_site_two_end (33 characters)
Site_three_huh_huh_huh_huh_huh_huh_huh_huh_site_three_end (49 characters)
Site_four_ooga_ooga_ooga_ooga_ooga_ooga_site_four_end (53 characters)
Site_five_whatsit_whatsit_whatsit_whatsit_whatsit_whatsit_site_five_end (65 characters)
Site_six_booga_booga_booga_booga_booga_booga_site_six_end (57 characters)
Suspect B
Site_one_yada_yada_yada_yada_site_one_end (41 characters)
Site_two_oy_oy_oy_oy_oy_oy_oy_oy_site_two_end (45 characters)
Site_three_huh_huh_huh_huh_huh_huh_huh_huh_site_three_end (57 characters)
Site_four_ooga_ooga_ooga_ooga_ooga_ooga_site_four_end (53 characters)
Site_five_whatsit_site_five_end (30 characters)
Site_six_booga_booga_booga_booga_site_six_end (45 characters)
The first PCR reaction makes copies of the DNA beginning with the sequence "site_one" through "site_one_end" and the other reactions proceed with "site_#" through "site_#_end." The DNA products are separated on a gel. The results appear something like this.
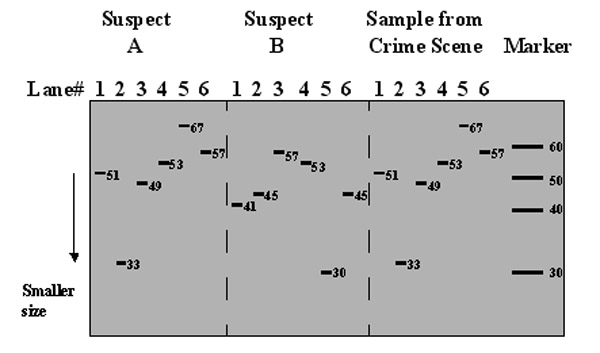
STR products separated on a gel extending the above analogy using sentence fragments.
In this case the sample from the crime scene matches suspect A.
Matching a sample to a known suspect is not the only way to find the source of the DNA evidence. The United States has a database of gene sequences known as CODIS. If the suspect is arrested in another crime that requires his or her DNA to be examined it can be matched. Or, if an evidence from another crime scene is taken, the DNA can be sequenced and entered into the database thus linking two crimes.
With high sensitivity and an excellent ability to discriminate among all but identical twins, is STR fingerprinting the perfect method? With 13 sites needing to be amplified contamination of multiple runs would be necessary for the results to lead to a false identification. However, contamination of a small portion of the runs could result in a missed identification.
A one in a trillion chance of matching doesn't mean a trillion to one odds that a person is guilty. Samples are still vulnerable to human errors and faults include mislabelling and evidence falsification. In regards to the latter, there are examples where suspects have gone out of their way to make sure a wrong sample is provided.
 Cross-section of a cell. The purple bodies are mitochondria. Cross-section of a cell. The purple bodies are mitochondria.
Mitochondrial DNA
Mitochondria are small bodies that exist inside of cells and provide the cell's energy. They have their own DNA and divide. The process of making spermatazoa eliminates mitochondria from the sperm cells, therefore all of our mitochondrial genes are inherited unchanged from the egg cells of our mothers. Mitochondrial DNA is small by comparison to nuclear DNA - only 16,000 "letters."
In the case of mtDNA, two highly variable regions are sequenced. This results in the entire area of DNA "spelled out." The forensic scientist then compares the suspect sample to the evidence sample. The analysis can be a bit complex, but when the sequences are identical they are determined to be a match. The FBI does not provide official statistics on how unlikely it is that two unrelated people would have the same type of mtDNA. The odds are probably greater than a thousand to one. If people are related through the same mother (or maternal grandmother and so on through the maternal line) they will have the same mtDNA.
There are several advantages and disadvantages to the use of mtDNA. Some samples can not be analyzed using STR or other nuclear DNA fingerprinting methods because they only have mtDNA. These may include hair shafts, bones and teeth. Also, since cells have multiple mitochondria, this method is better able to find DNA in minute or highly degraded samples. The disadvantages are the lack of resolution among related individuals and the smaller number of mtDNA samples entered into the SORIS database. While some think of mtDNA as the method used when only hair samples are available, the root of hairs have nuclear DNA and STR fingerprinting is also an option.
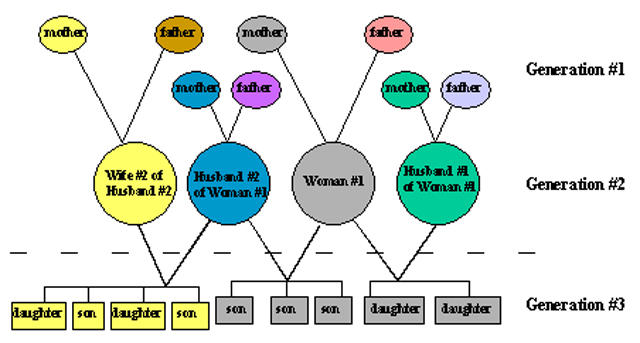
MtDNA inheritance across three generations.
Individuals with the same colored circles share the same mtDNA.
Although many have the same mtDNA, even maternal half-siblings,
there are still eight mtDNA types represented among these 21 individuals.
[Continued in Part Three. Findings from the West Memphis Case.] |







![]()
![]()